import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
def ppm_to_da(error_ppm, theoretical_mass):
error_da = theoretical_mass / ((1/ error_ppm) * 1000000)
return error_da
def flatten(l):
return [item for sublist in l for item in sublist]
def generate_df(masses, errors):
mass_error = pd.DataFrame(columns=["Mass", "Error (ppm)", "Error (Da)"])
mass_error["Mass"] = masses * len(errors)
mass_error["Error (ppm)"] = flatten([[e] * len(masses) for e in errors])
mass_error["Error (Da)"] = mass_error.apply(
lambda row: ppm_to_da(row["Error (ppm)"], row["Mass"]),
axis=1
)
return mass_error
masses = [400, 1200]
errors = [25, 50, 100]
mass_error = generate_df(masses, errors)
sns.lineplot(data=mass_error, x="Mass", y="Error (Da)", hue="Error (ppm)")
plt.show()
Last active
October 23, 2025 06:07
-
-
Save RalfG/a74448fad9a0766a8e5afbbe3a6541d7 to your computer and use it in GitHub Desktop.
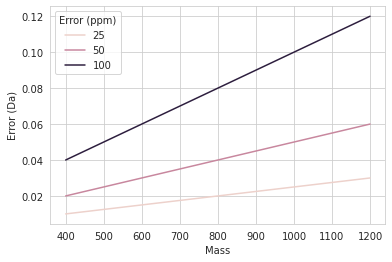
Dalton vs ppm mass error in mass spectrometry
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Awesome! Just love the colab for teaching, so here is a copy https://colab.research.google.com/drive/1Y55cjN3HahxInkwCOjQwNG_NXEZ0xmKl?usp=sharing 🙏