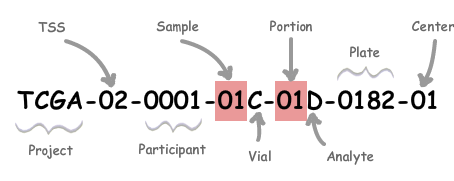
There are many instances where more than one aliquot is provided by TCGAbiolinks datasets, but you will only need one of those. In this case, GDC has offered a standard set of Replicate Sample rules to select the most 'scientifically advantageous' aliquot for study.
From the GDC Documentation Encyclopedia
| Label | Identifier for | Value | Value Description | Possible Values |
|---|---|---|---|---|
| Analyte | Molecular type of analyte for analysis | D | The analyte is a DNA sample | See Code Tables Report |
| Plate | Order of plate in a sequence of 96-well plates | 182 | The 182nd plate | 4-digit alphanumeric value |
| Portion | Order of portion in a sequence of 100 - 120 mg sample portions | 1 | The first portion of the sample | 01-99 |
| Vial | Order of sample in a sequence of samples | C | The third vial | A to Z |
| Project | Project name | TCGA | TCGA project | TCGA |
| Sample | Sample type | 1 | A solid tumor | Tumor types range from 01 - 09, normal types from 10 - 19 and control samples from 20 - 29. See Code Tables Report for a complete list of sample codes |
| Center | Sequencing or characterization center that will receive the aliquot for analysis | 1 | The Broad Institute GCC | See Code Tables Report |
| Participant | Study participant | 1 | The first participant from MD Anderson for GBM study | Any alpha-numeric value |
| TSS | Tissue source site | 2 | GBM (brain tumor) sample from MD Anderson | See Code Tables Report |
Portion/analyte From GDC Documentation Encyclopedia
| Code | Definition |
|---|---|
| D | DNA |
| G | Whole Genome Amplification (WGA) produced using GenomePlex (Rubicon) DNA |
| H | mirVana RNA (Allprep DNA) produced by hybrid protocol |
| R | RNA |
| T | Total RNA |
| W | Whole Genome Amplification (WGA) produced using Repli-G (Qiagen) DNA |
| X | Whole Genome Amplification (WGA) produced using Repli-G X (Qiagen) DNA (2nd Reaction) |
Natively, GDC uses two filters to only one sample per patient in the GDC Firehose repository, we shall follow the same set of rules.
Analyte Replicate Filter
If the aliquot is an RNA sample: H > R > T
somewhat arbitrary and subject to change, since it is not clear at present whether H or R is the better protocol
If there are still technical replicates left: max(plate number)
If aliquot is a DNA sample: max(plate number)
(D > G / W / X)
If there are still replicate left, use
Sort Replicate Filter
chooses the aliquot with the highest lexicographical sort value
Examples of the Sort Replicate Filter
| removed | TCGA-A6-2677-01A-01D-A274-01 |
| chosen | TCGA-A6-2677-01B-02D-A274-01 |
| removed | TCGA-A6-2684-01A-01D-A274-01 |
| chosen | TCGA-A6-2684-01C-08D-A274-01 |
| removed | TCGA-A6-6650-01A-11D-A274-01 |
| chosen | TCGA-A6-6650-01B-02D-A274-01 |
| removed | TCGA-06-0138-01A-01D-0236-01 |
| chosen | TCGA-06-0138-01A-02D-0236-01 |
| removed | TCGA-06-0211-01A-01R-1849-01 |
| chosen | TCGA-06-0211-01B-01R-1849-01 |
The whole list of replicate filtering example can be found in http://gdac.broadinstitute.org/runs/stddata__2014_01_15/samples_report/filteredSamples.2014_01_15__00_00_11.txt